Dr Heba Sailem Sir Henry Wellcome Research Fellow, Institute of Biomedical Engineering. Pic supplied
AI speeds up identification of the role of colon cancer gene functions
Researchers in Oxford and Zurich have used artificial intelligence (AI) and computer vision technology to detect genes involved in changing the organisation of colon cells which is a critical step in cancer development. This technology resulted in the discovery of a role for smell sensing genes in this process. The technology played an important role making the research speedier and more efficient, automating a process that would have previously been done manually.
The new study, led by Dr Heba Sailem, Sir Henry Wellcome Research Fellow at the Institute of Biomedical Engineering in collaboration with Professor Jens Rittscher and University of Zurich’s Lucas Pelkmans, has found that patients whose colon cancer cells show the ‘expression’ of certain smell-sensing genes are more likely to have worse outcomes, especially those with more severe cases of cancer.
Humans have around 400 ‘smell-sensing’ genes which activate in a combination of ways to allow us to smell the ranges of smells that we do. However, the genes have been found to be expressed in parts of the body other than the nose, with their role previously remaining a mystery.
The ‘expression’ of a gene is when the information that is stored in DNA is translated into instructions for making proteins or other molecules. Gene expression can act as an on/off switch to control when proteins are made and how many. So, the expression of these smell-sensing genes means that the instructions for these particular genes are in use. The levels of the genes can be reduced by experimental techniques called “perturbations” to study the role of the gene in the cell.
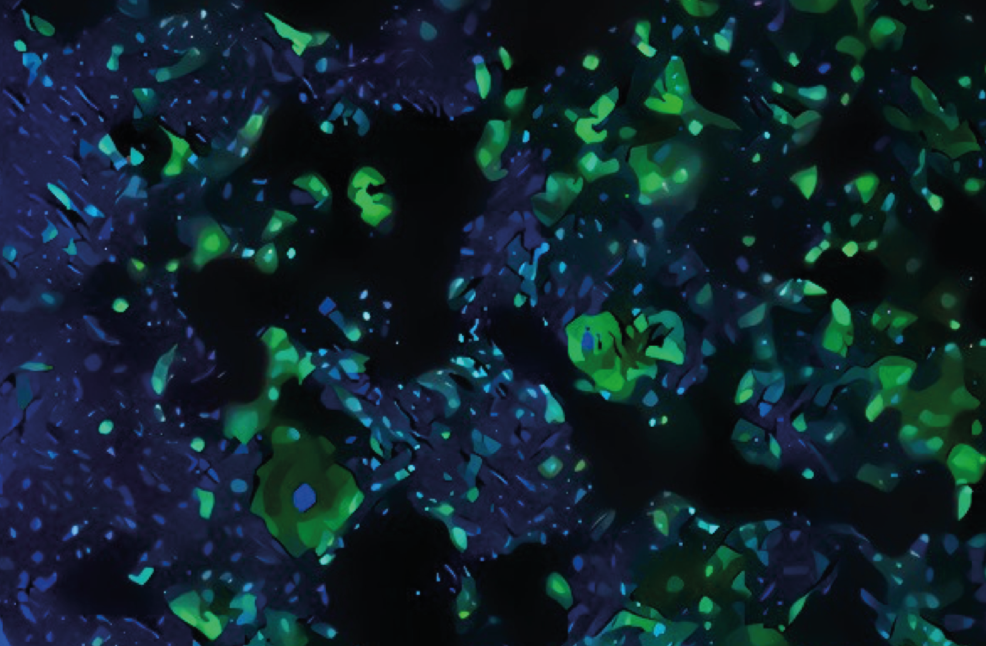
Colon cancer cells visualisation. Pic: Dr Heba Sailem
When researching the development of cancer, a key thing to look at is the organisation of cells in body tissue. Dr Sailem explains: “Cancer is often characterised with the loss of tissue structure which can be driven by certain gene alterations or stresses. It is crucial to understand which genes play a role in this process to be able to develop therapies that target cancer development.”
The researchers used multiple layers of Artificial Intelligence including a computer vision algorithm to detect changes in cell appearance and organisation. The algorithm was fed information from robotic microscopy, in collaboration with researchers from the University of Zurich, to image millions of colon cancer cells.
Following the perturbation (or decrease in the expression) of every gene in each individual colon cancer cell, the study found that smell-sensing genes are strongly associated with how cells spread and align with each other. Reducing the expression of smell-sensing genes can inhibit cells from spreading, potentially by restraining the ability of cells to move. The same behaviour is also observed in the perturbation of key cancer genes. In contrast, having higher levels of these genes might increase cell motility.
Dr Sailem adds: “It is like activating a sixth sense that allows cancer cells to smell their way outside the toxic tumour environment which can result in spreading cancer to other parts of the body and make things worse for the patient.”
Artificial intelligence was crucial in accelerating the speed and efficiency of this research. The computer is trained using an encyclopaedia of knowledge on the functions of genes collected over decades and automates the process of identifying cell patterns in the images. Previously, this type of research would have relied on the slower and more costly method of human experts identifying examples of changes in cell appearance.
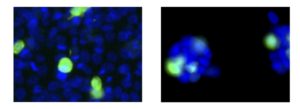
Colon cancer cells before (left) and after (right) the perturbation of a gene. Pic: Dr Heba Sailem
“Using the developed AI system, we can now learn much more from these experiments and accelerate the identification of genes that alter the structure of tissues in cancer.”, Sailem says. Gene editing technology CRIPSR (Clustered Regularly Interspaced Short Palindromic Repeats) is now routinely used to reduce the level of every gene in the cell out of around 20,000 genes to study how changing the level of the gene affects cancer cell behaviour.
Parallel with advances in gene editing technologies, such as CRISPR, this research could enable new avenues in identifying the functions of genes in different cancer types which is vital for cancer therapy and understanding cancer evolution.
‘KCML: a machine‐learning framework for inference of multi‐scale gene functions from genetic perturbation screens,’ is published in Molecular Systems Biology.
This article is an edited version of this news story by University of Oxford Department of Engineering Science.
